ENMTools tests can take a long time to run, due to the number of Maxent runs necessary to construct a distribution of expected overlaps. For that reason, people occasionally ask whether there's a way to break up analyses and run them on a cluster, or just manually spread them across several computers. People also occasionally ask whether it's possible to use ENMTools with non-Maxent methods of ENM construction. There's no built-in way to do either of these at present, and given the number of different ways that people might want to do this I'm not sure there will be. However, it CAN be done! It requires a bit of extra work, but it's not too bad, particularly with a few simple new tools that I'm presenting here.
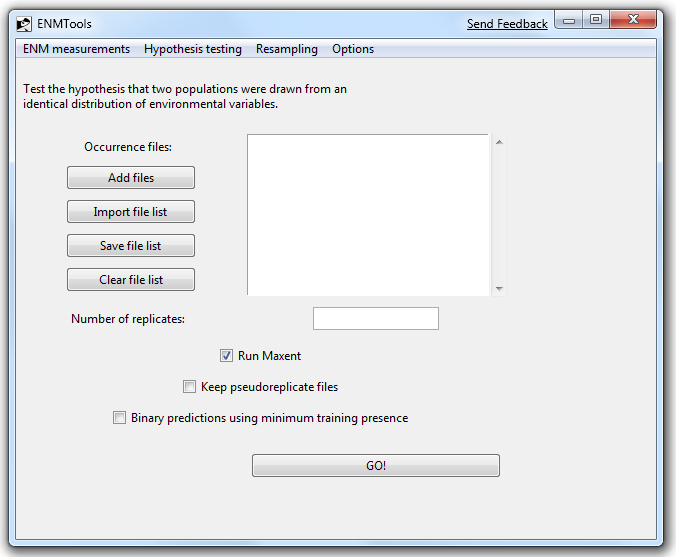
Let's say you want to do an identity test, and split it up so that replicates are submitted to a cluster. For starters, we'll just set up the test in ENMTools as usual but de-check the box marked "Run Maxent". This will cause ENMTools to generate the data sets necessary for the test, but the data will not be sent to Maxent to analyze.
Okay, now we have a file that has a whole bunch of replicates in it. What we would like to do is analyze each of those separately. I've got a really simple no-frills script
here that simplifies this process. All you need to do is drop this file into the directory where your file of replicates is, then go to a command prompt in that directory. Type "splitcsv.pl INFILE", where INFILE is the name of your .csv file of replicates. In short order, you will have a new .csv file in that directory for each replicate in the input file. Note: you can also use this little tool to split a large file up by species. It will spit out a file for each unique species name in the input file. Another note:
this script will overwrite files without asking, so it's best to run it in its own directory!At this point it's up to you to figure out how to submit jobs to your cluster using the Maxent command line options. One thing to keep in mind: Maxent by default writes to a file called maxentResults.csv. If you've got a shared file system, or just a directory on a single multi-core computer that's being shared by two different simultaneous Maxent jobs, both of those jobs will by default try to write to the same results file. Java will happily allow multiple instances of Maxent to fight over the same filehandle, but what comes out in that results file will be incomplete and quite possibly not useful. I've just set my stuff up so that it writes to different directories, but it's possible that using the "perspeciesresults" option will fix this issue as well. On a side note: if you're doing jackknife or bootstrap on a cluster, you pretty much can't have multiple instances of Maxent writing to the same directory. The perspeciesresults option doesn't work for resampling, and having multiple instances writing to the same output file simultaneously will cause Maxent to fail when calculating summary grids. Just FYI.
All right, now you've gotten all of your runs finished and want to bring your data together to construct a distribution of overlaps. This is where a handy new half-developed tool in ENMTools becomes useful: scripting mode. You'll need
the newest test version of ENMTools for this. I'll admit that it's a little weird to have a scripting interface for a program that is itself just an elaborate scripting interface for Maxent, but that doesn't mean it's not useful. At present the scripting interface is only hooked up for a few functions, and I'm not even close to writing a comprehensive manual entry for it. However, it works for our purposes.
What you need to do is build a .csv file that has a line for each comparison to be made. The script command is "measureOverlap". Capitalization is not important. A typical line looks like this:
measureOverlap,c:/sample data/species1_rep1.asc,c:/sample data/species2_rep1.asc,testing_measureOverlap1
The first entry is the command, the next two are the two files to compare, and the last entry is the name for the analysis. If you're doing a comparison between two species using 100 replicates, you need 100 lines in your script file, each with its own name for generating output files. In cases like this, the "concatenate" function in Excel is your best friend. Once you've got your script file, just go to Options->Run Script File in ENMTools and let it do its thing.
Now you've got two output files per replicate (I and D), and you'd really like to have all of those in one file. In Windows, the easy way to do this is to go to a command prompt and use the "copy" command with some clever wildcards. In the above example, let's say I've got 100 files named "testing_measureOverlap1_I_output.csv" to "testing_measureOverlap100_I_output.csv", and likewise for D. The appropriate command would be something like:
copy testing_measureOverlap*_I_output.csv collected_I_scores.csv
This would concatenate all of those output files for I into one csv file that can then be edited in Excel. There's going to be a bit of cleanup to do, since each of those files had its own header line and two copies of each score. That's all fairly easy, though, and should only take a couple of minutes with some clever sorting.
Everything I've said here goes for other modeling methods as well - you can generate data sets and analyze them in whatever software you like, and then use the scripting interface to build your distributions. When I get a chance I plan to make an interface that will make the comparison of multiple files easier (i.e., send everything to one formatted outfile). Seeing as I'm currently in a blind rush to finish my dissertation before my postdoc starts, though, I don't think that's going to happen too soon.